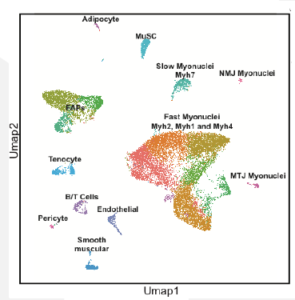
Regulation of the super enhancer at the fast Myosin heavy chain locus
The overall goal of this project is to define the regulatory mechanisms determining the myofiber type of adult skeletal muscle and more specifically the mechanisms controlling the expression of a single myosin heavy chain (Myh) gene in the hundreds of nuclei present in a given fiber. The major myofiber types are generally defined by the Myh genes that they express. However, we lack a comprehension of the regulatory mechanisms that determine which Myh gene will be expressed within a myofiber. The fast Myh (fMyh) locus in mammals is composed of Myh2, Myh1, Myh4, Myh8 and Myh13 genes arranged in 350Kb. Previous work from our group has identified by 4C experiments a 42kb cis-regulatory module (CRM) upstream of Myh2 that can interact with the promoters of Myh2 in the slow soleus and with the promoter of Myh4 in the fast quadriceps. This CRM is composed of discrete DNA elements as revealed by single nucleus ATAC-seq (snATAC-seq) experiments performed with adult skeletal muscles and we have shown that this CRM acts as a super-enhancer (SE) controlling fast Myh genes expression. We demonstrated that innervation is required to coordinate fMyh gene expression in the hundreds of myonuclei present in an adult myofiber.
The research proposed will allow to fully characterize how the fast subtype myofiber gene expression program is established. In particular, we will test how the SE at the Myh locus is responsible for organizing alternate DNA conformations to ensure that only a single Myh gene is expressed in the muscle fiber. SnRNA-seq and snATAC-seq experiments will be performed with adult muscles of wt and PMA (peroneal muscular atrophy animals whose distal hindlimb muscles have never been innervated). These experiments will identify the transcription factors and signaling pathways linking fast motoneuron firing, SE activity and myonuclei coordination. Identified DNA regions relaying fast motoneuron subtypes will be validated by transient transfection experiments in adult muscles.
Publications de l’équipe relatives au projet de stage
Dos Santos,M., Backer,S., Auradé,F., Man-Kin Wong,M., Wurmser,M., Pierre,R., Langa,F., Do Cruzeiro,M., Schmitt,A., Concordet,J-P., Dilworth,F.J., Noordermeer,D., Sotiropoulos,A., Relaix,F., Sakakibara,I., Maire,P. A fast Myh super enhancer dictates adult muscle fiber phenotype through competitive interactions with the fast Myh genes. 2021. bioRxiv 2021.04.17.438406; doi: https://doi.org/10.1101/2021.04.17.438406
Dos Santos,M.,et al. 2020. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat Commun 11, 5102. https://doi.org/10.1038/s41467-020-18789-8
Wurmser,M.,et al. 2020. SIX1 and SIX4 homeoproteins regulate PAX7+ progenitor cell properties during fetal epaxial myogenesis. Development. 147(19):dev185975. doi: 10.1242/dev.185975.
Randrianarison-Huetz, V. et al. 2018. Srf controls satellite cell fusion through the maintenance of actin architecture. J.Cell.Biol. 217(2):685-700.
Santolini, M.*,et al. 2016. MyoD reprogramming requires Six1 and Six4 homeoproteins: genome-wide cis-regulatory module analysis. Nucleic Acids Res. 44(18):8621-8640.
Sakakibara,I., et al 2014. Six homeoproteins and a linc-RNA cooperate at the fast MYH locus to lock terminal fast myofibre phenotype. PLoS Genet. 10 (5) : e1004386